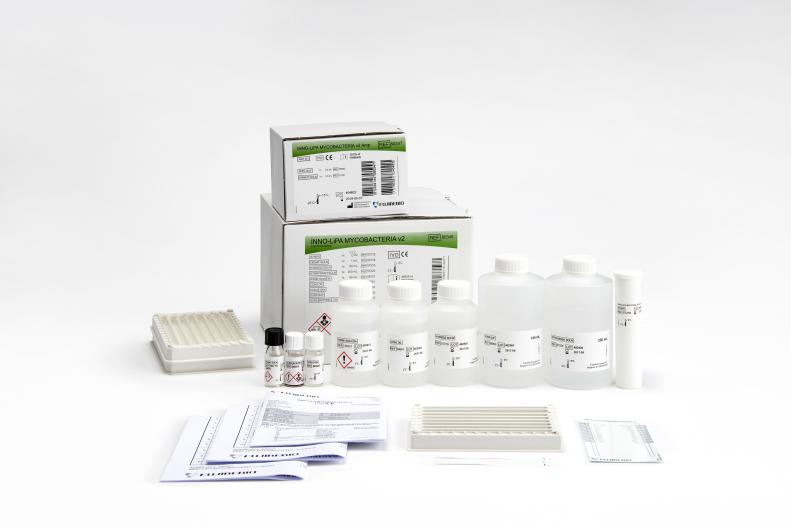
INNO-LiPA® MYCOBACTERIA v2
One strip – one test for the detection of the genus Mycobacterium and 16 mycobacterial species.
INNO-LiPA MYCOBACTERIA v2 is a line probe assay for the simultaneous detection and identification of the genus Mycobacterium and 16 different mycobacterial species.
The test is based on the nucleotide differences in the 16S-23S rRNA spacer region and can be performed starting from either liquid or solid culture.
The following Mycobacterium species can be detected simultaneously: M. tuberculosis complex, M. kansasii, M. xenopi, M. gordonae, M. genavense, M. simiae, M. marinum and M. ulcerans, M. celatum, MAIS, M. avium, M. intracellulare, M. scrofulaceum, M. malmoense, M. haemophilum, M. chelonae complex, M. fortuitum complex, and M. smegmatis.
Product number 80346
Product number 80347